Notice
Recent Posts
Recent Comments
Link
| 일 | 월 | 화 | 수 | 목 | 금 | 토 |
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| 8 | 9 | 10 | 11 | 12 | 13 | 14 |
| 15 | 16 | 17 | 18 | 19 | 20 | 21 |
| 22 | 23 | 24 | 25 | 26 | 27 | 28 |
| 29 | 30 |
Tags
- TRANSFORMER
- Super-Resolution
- Zero-Shot sr
- arbitrary scale
- SegFormer
- SISR
- DIINN
- hypergraph
- Cell-tissue
- FRFSR
- GNN
- GNN in image
- Cityscapes
- Cell detection
- implicit representation
- session-based recommendation
- Graph
- ADE20K
- deep learning
- CrossNet
- Feature reuse
- LIIF
- INR
- Referense Super Resoltuion
- Tissue segmentation
- Reference Super-Resolution
- graph neural network
- ConvNeXt
- TAAM
- Reference-based SR
Archives
- Today
- Total
딥러닝 분석가 가리
EMCAD: Efficient Multi-scale Convolutional Attention Decoding for Medical Image Segmentation 본문
딥러닝 논문 리뷰
EMCAD: Efficient Multi-scale Convolutional Attention Decoding for Medical Image Segmentation
AI가리 2024. 7. 16. 15:08EMCAD: Efficient Multi-scale Convolutional Attention Decoding
for Medical Image Segmentation (CVPR 2024)
Abstract
- In medical image segmentation, an efficient and effective decoding mechanism is crucial
- These decoding mechanisms usually come with high computational costs
- EMCAD, a new efficient multi-scale convolutional attention decoder, designed to optimize both performance and computational efficiency
- Leverages a unique multi-scale depth-wise convolution block
- Employs channel, spatial, grouped(large-kernel) gated attention mechanisms
- 1.91M parameters and 0,381G FLOPs, when using a standard encoder
- Across 12 datasets, belonging to six medical image segmentation tasks, EMCAD achieves state-of-the-art (SOTA) performance with 79.4% and 80.3% reduction in #Params and # FLOPs, respectively
- EMCAD's adaptability to different encoders and versatility across segmentation task
Introduction
- Automated segmentation of medical images is crucial, as it classifies pixels to identify critical regions such as lesions, tumors, or entire organs
- A variety of U-shaped CNN architectures have become standard techniques for this purpose
- Attention mechanisms have also been integrated into these models to enhance feature maps and improve pixel-level classification
- Vision transformers have shown promise in medical image segmentation tasks by capturing long-range dependencies among pixels through self-attention(SA) mechanisms
- Hierarchical vision transformers have been introduced to improve the performance in this field further
- While the SA excels at capturing global information, it is less adept at understanding the local spatial context
- Nevertheless, these methods can still be computationally demanding because they frequently employ costly convolutional blocks
- To address the limitations above, the authors introduce EMCAD, and efficient multi-scale convolutional attention decoding using a new multi-scale depth-wise convolution block
- EMCAD enhances the feature maps via efficient multi-scale convolutions while incorporating complex spatial relationships and local attention through the use of channel, spatial, and grouped (large-kernel) gated attention mechanisms
Contributions
- New Efficient Multi-scale Convolutional Decoder
- Efficient Multi-scale Convolutional Attention Module
- Large-kernel Grouped Attention Gate
- Improved Performance
Methodology

Efficient multi-scale convolutional attention decoding (EMCAD)
- Introduce EMCAD to process the multi-stage features extracted from pre-trained hierarchical vision encoders for high-resolution semantic segmentation
- EMCAD consists of efficient multi-scale convolutional attention modules (MSCAMs), large-kernel grouped attention gates (LGAGs), efficient up-convolution blocks (EUCBs), and segmentation heads (SHs)
Large-kernel grouped attention gate (LGAG)
- Introduce LGAG to progressively combine feature maps with attention coefficients, which are learned by the network to allow higher activation of relevant features and suppression of irrelevant ones
- Employ a gating signal derived from higher-level features to control the flow of information across different stages of the network, thus enhancing its precision for medical image segmentation
- Upsampled features and features from skip connections are inputted, and the upsampled feature is multiplicated
- Due to using a 3×3 kernel, instead of a 1×1 kernel, group convolutions, LGAG captures comparatively larger spatial contexts with less computational cost
Multi-scale convolutional attention module (MSCAM)
- MSCAM is an efficient multi-scale convolutional attention module to refine the feature maps
- MSCAM consists of a channel attention block (CAB), a spatial attention block (SAB), and an efficient multi-scale convolution block (MSCB)
- Using depth-wise convolution in multiple scales, MSCAM is more effective with significantly lower computational cost
Multi-scale convolution block (MSCB)
- To enhance the features generated by our cascaded expanding path
- Follow the design of the inverted residual block (IRB) of MobileNetV2
- However, unlike IRB, MSCB performs depth-wise convolution at multiple scales and uses channel_shuffle to shuffle channels across groups (to incorporate relationships among channels)
- Use multi-scale depth-wise convolution (MSDC), the recursively updated input x, where the input x is residually connected to the previous depth-wise convolution block
- Residually connected for better regularization
Channel attention block (CAB)
- Use channel attention block to assign different levels of importance to each channel, thus emphasizing more relevant features while suppressing useful ones
- Apply the adaptive maximum pooling and adaptive average pooling to the spatial dimensions to extract the most significant feature of the entire feature map channel
- Reduce the number of channels r = 1/16 times separately using a point-wise convolution and recover the original channels using another point-wise convolution
- Use the Sigmoid function and Hadamard product for channel attention
Spatial attention block (SAB)
- To mimic the attentional processes of the human brain by focusing on specific parts of an input image
- SAB determines where to focus in a feature map; then it enhances those features
- Enhance the model's ability to recognize and respond to relevant spatial features, which is crucial for image segmentation
- Pool maximum and average values along the channel dimension to pay attention to local features
- Use a large kernel 7×7 convolution layer to enhance local contextual relationships among feature
- Use the Sigmoid function and Hadamard product for spatial attention
Efficient up-convolution block (EUCB)
- Use an EUCB to progressively upsample the feature maps of the current stage to match the dimension and resolution of the feature maps from the next skip connection
- Use UpSampling with scale factor 2, 3×3 depth-wise convolution for enhance feature maps followed BN and ReLU, and finally 1×1 kernel convolution layer to reduce the #channels
Segmentation head (SH)
- Use segmentation heads to produce the segmentation outputs from the refined feature maps of the four stages of the decoder
- Use 1×1 kernel convolution to refine feature maps, output with #channel equal to #classes
Overall architecture
- To show the generalization, effectiveness, and ability to process multi-scale features for medical image segmentation, the authors integrate our EMCAD decoder alongside tiny(PVTv2-B0) and standard (PVTv2-B2) networks of PVTv2
- To adopt PVTv2, the authors first extract the features (X1, X2, X3, and X4) from four layers and feed them into EMCAD, then process them and produce four segmentation maps that correspond to the four stages of the encoder network
Multi-stage loss and outputs aggregation
Loss aggregation
- Adopt a combinatorial approach to loss combination called MUTATION, inspired by the work of MERIT
- This involves calculating the loss for all possible combinations of predictions derived from 4 heads, a total of 15 unique predictions, and then summing these losses
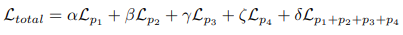
Output segmentation maps aggregation
- Sigmoid function for binary or Softmax function for multi-class segmentation
Experiments
Dataset (Split 8:1:1 in ClinicDB, Kvasir, ColonDB, ETIS, BKAI, ISIC18, DSB18, EM, and BUSI datasets)
- Polyp segmentation
- Kvasir(1000 images), ClinicDB(612 images), ColonDB(379 images), ETIS(196 images), BKAI(1000 images)
- These datasets contain images from different imaging centers/clinics, having greater diversity in image nature as well as the size and shape of polyps
- Synapse multi-organ dataset
- Abdominal CT scans (18 scans for training, 12 scans for validation)
- gallbladder (GB), left kidney (KL), right kidney (KR), liver, pancreas (PC), spleen (SP), and stomach (SM)
- ACDC dataset
- Cardiac MRI scans (70 for training, 10 for validation, 20 for testing)
- right ventricle (RV), myocardium (Myo), left ventricle (LV)
- Skin lesion segmentation
- ISIC17 (2000 training, 150 validation, and 600 testing images), ISIC18 (2594 images)
- Breast cancer segmentation
- BUSI (437 benign and 210 malignant)
- Cell nuclei/structure segmentation
- DSB18 (670 images), EM (30 images)
Evaluation metrics
- DICE score and HD95
Implementation details
- Pytorch 1.11.0 on a single NVIDIA RTX A6000 GPU
- Utilize ImageNet pre-trained PVTv2-B0 and PVTv2-B2 as encoders
- MSDC kernel [1, 3, 5] through an ablation study
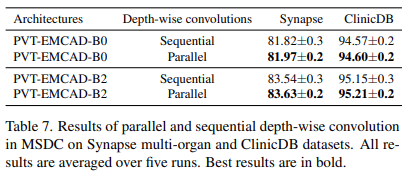
- Use the parallel arrangement of depth-wise convolutions in all experiments
- AdamW optimizer with learning rate and weight decay of 1e-4
- Epochs 200, batch size 16, Synapse (300, 6), ACDC (400, 12)
- Image size 352×352 and use a multi-scale {0.75, 1.0, 1.25} training strategy
- 256×256 for BUSI, EM, and DSB18
- 224×224 for Synapse and ACDC
- Loss function = 0.3 Cross-entropy loss + 0.7 DICE loss
- For binary segmentation use binary cross-entropy and weight IoU loss function
Results
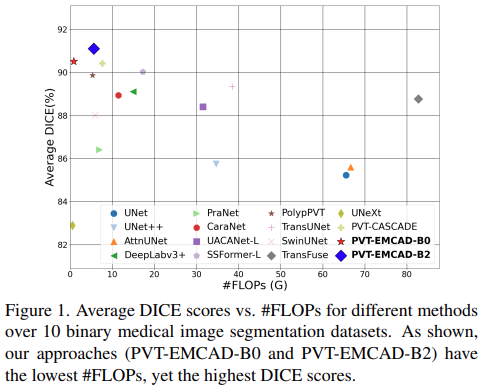
Results of binary medical image segmentation
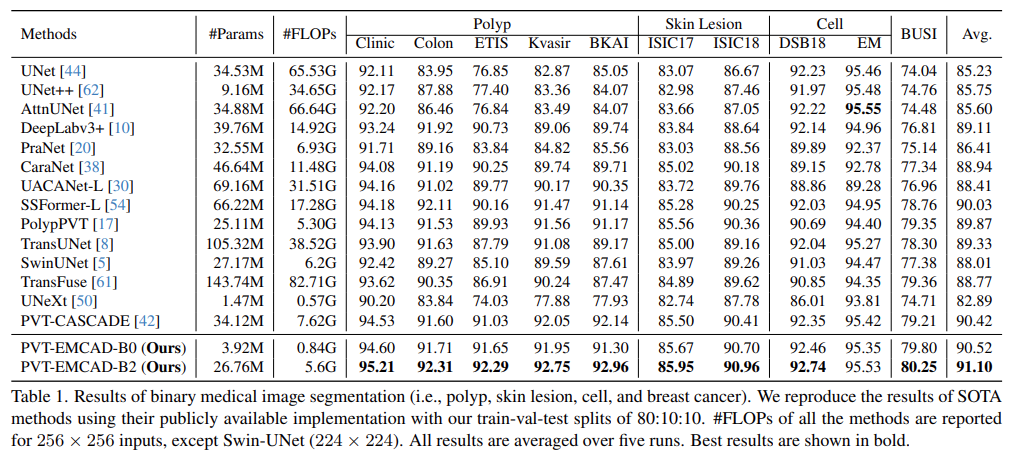
- PVT-EMCAD-B2 attains the highest average DICE score (91.10%) with only 26.76M parameters and 5.6G FLOPs
- The multi-scale depth-wise convolution in the EMCAD decoder, combined with the transformer encoder, contributes to these performance gains
Polyp, Skin lesion, Cell, Breast cancer segmentation
- PVT-EMCAD-B2 achieves the new SOTA results in these segmentation datasets
- On the EM, PVT-EMCAD-B2 secures the second-best DICE score, offering significantly lower computational costs
Results of abdomen organ segmentation
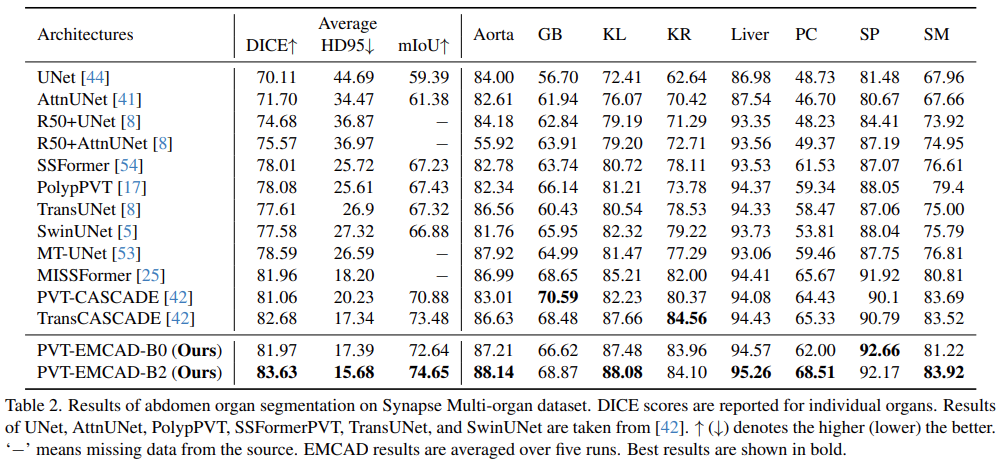
- PVT-EMCAD-B2 achieves the highest average DICE score and HD95
- EMCAD decoder boosts individual organ segmentation, significantly outperforming SOTA methods on six of eight organs
Results of cardiac organ segmentation
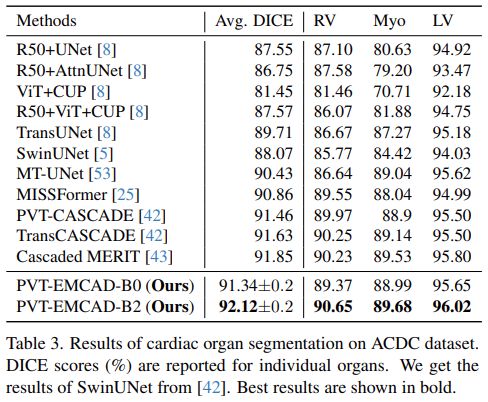
- PVT-EMCAD-B2 achieves the highest average DICE score of 92.12%, besides, it has better DICE scores in all three organ segmentation
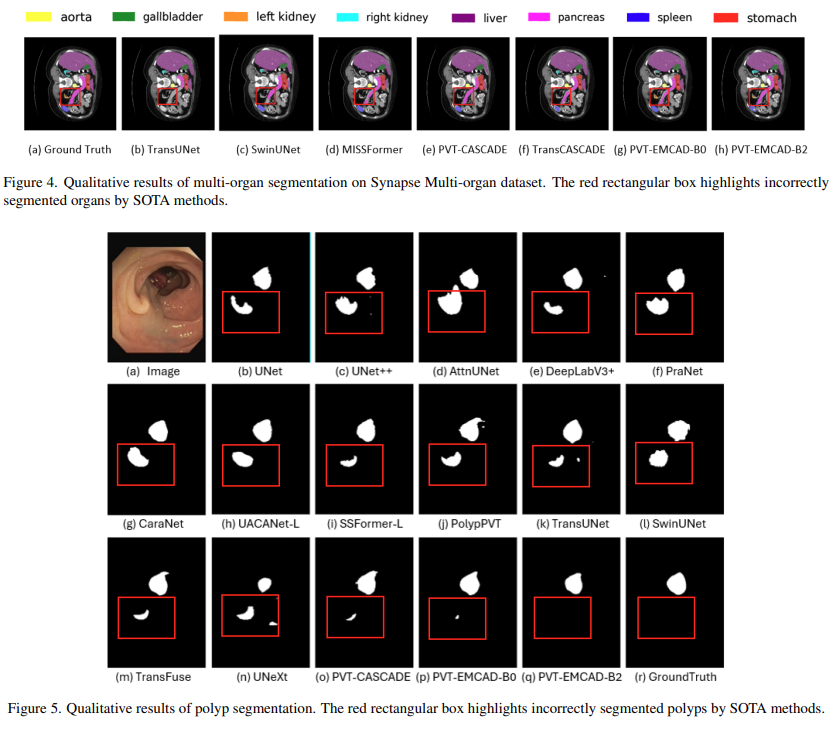
Qualitative results
Ablation Studies
Effect of different components of EMCAD
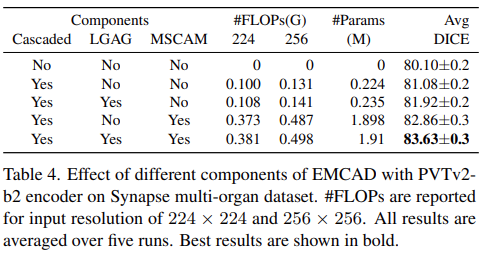
- Conduct a set of experiments on the Synapse multi-organ dataset to understand the effect of different components of EMCAD
- Cascaded, LGAG, MSCAM helps to improve performance
Effect of multi-scale kernels in MSCAM
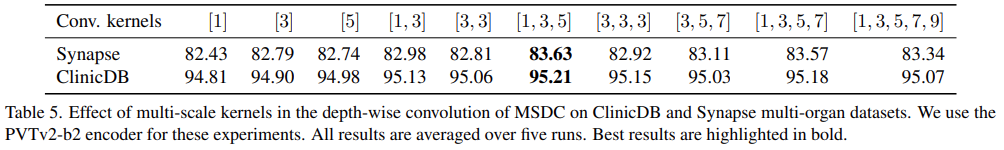
- Conducted another set of experiments on Synapse and ClinicDB to understand the effect of different multi-scale kernels used for depth-wise convolution in MSDC
- [1, 3, 5] kernels is best
Comparison with the baseline decoder
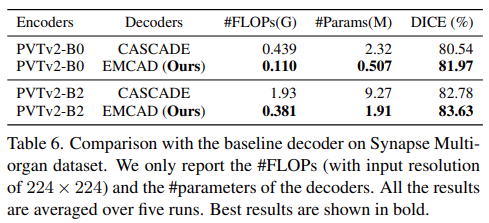
- Report the experimental results with the computational complexity of EMCAD and baseline decoder namely CASCADE
Parallel vs. sequential depth-wise convolution
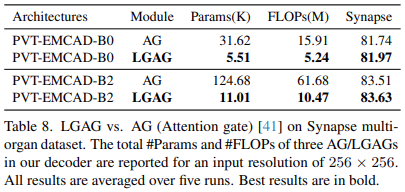
Effectiveness of LGAG over attention gate
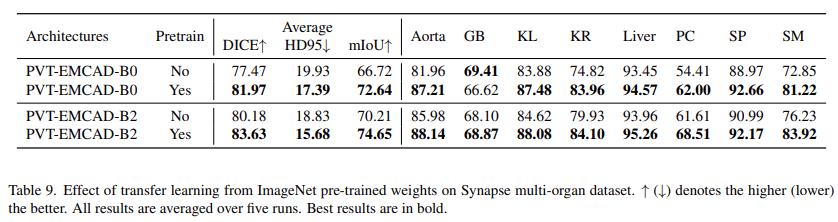
Effect of transfer learning from ImageNet pre-trained weights
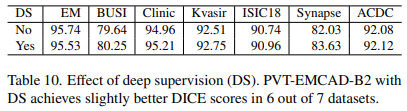
Effect of deep supervision
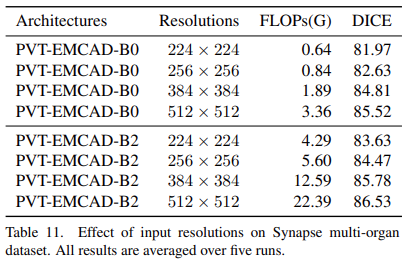
Effect of input resolutions
Conclusion
- Presented EMCAD, a new and efficient multi-scale convolutional attention decoder designed for multi-stage feature aggregation and refinement in medical image segmentation
- EMCAD employs a multi-scale depth-wise convolution block, which is key for capturing diverse scale information within feature maps, a critical factor for precision in medical image segmentation
- This method achieved SOTA
- The authors anticipate that the EMCAD decoder will be a valuable asset in enhancing a variety of medical image segmentation and semantic segmentation tasks
Paper
'딥러닝 논문 리뷰' 카테고리의 다른 글
| Learning with Unmasked Tokens DrivesStronger Vision Learners (2) | 2024.09.09 |
|---|---|
| Self-Supervised Hypergraph Convolutional Networks for Session-based Recommendation Review (0) | 2024.04.27 |
| NestedFormer Review (0) | 2024.04.03 |
| ConvNeXt : A ConvNet for the 2020s (0) | 2024.01.16 |
| Single Image Super-Resolution via a Dual Interactive Implicit Neural Network Review (0) | 2023.11.03 |
Comments




